Description
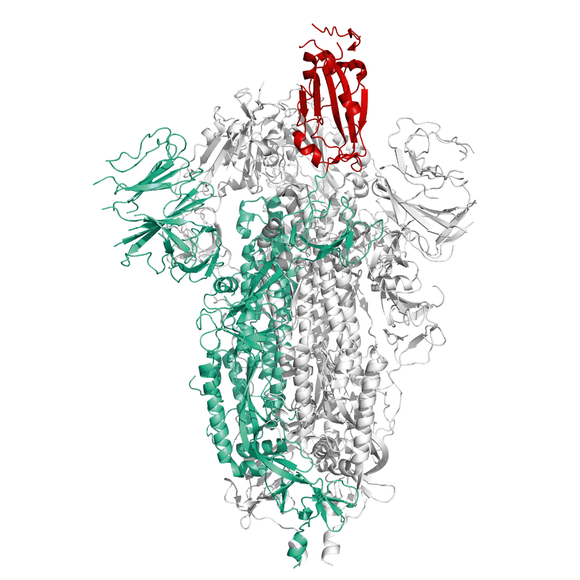
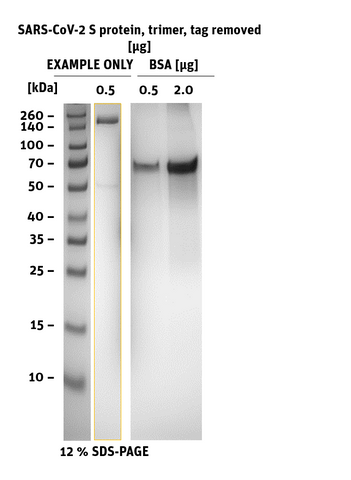
Recombinant Spike protein of SARS-CoV-2 (COVID-19) from Wuhan pneumonia virus as a stabilized trimer with deletion of the internal Furin-site, C-terminal Strep- and His-Tag removed. The S protein plays key parts in the induction of neutralizing-antibody and T-cell responses, as well as protective immunity.
- Product Name: SARS-CoV-2 (COVID-19) S Protein, Tag-removed stabilized trimer
- Catalog No.: P2020-029
- RefSeq Links: NC_045512.2; MN908947.3; YP_009724390.1; QHD43416.1; GeneID: 43740568; UniProt: P0DTC2
- Synonyms: SARS-CoV-2; coronavirus; SARS-CoV-2 spike protein; S glycoprotein; spike glycoprotein; 2019-nCoV; COVID-2019; COVID-19